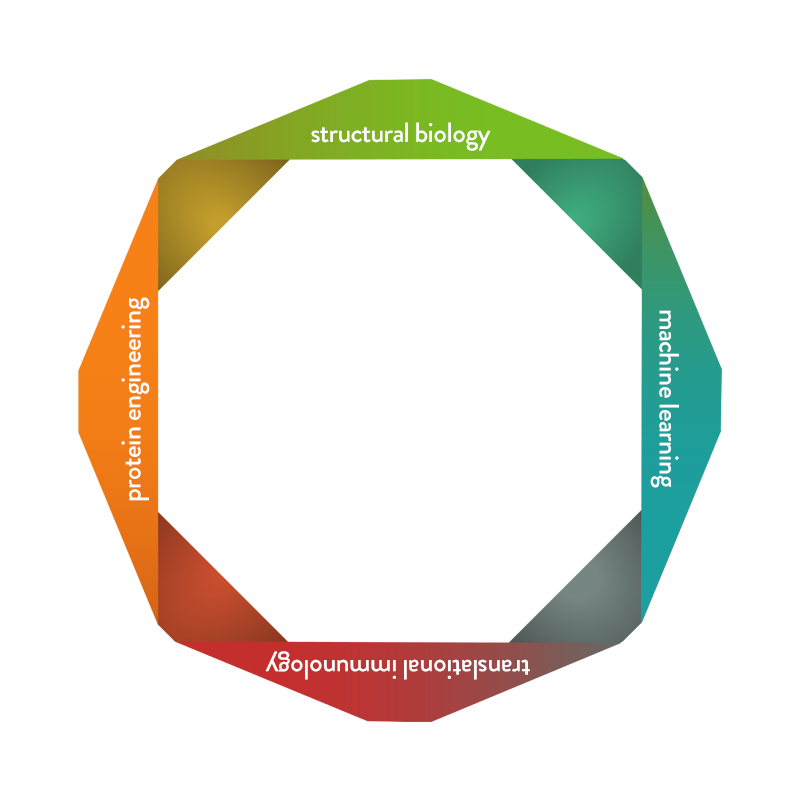
What We Do
Our trailblazing team generates state-of-the-art proprietary machine learning algorithms and statistical models to systematically analyze and modify protein sequences.
Where It Leads
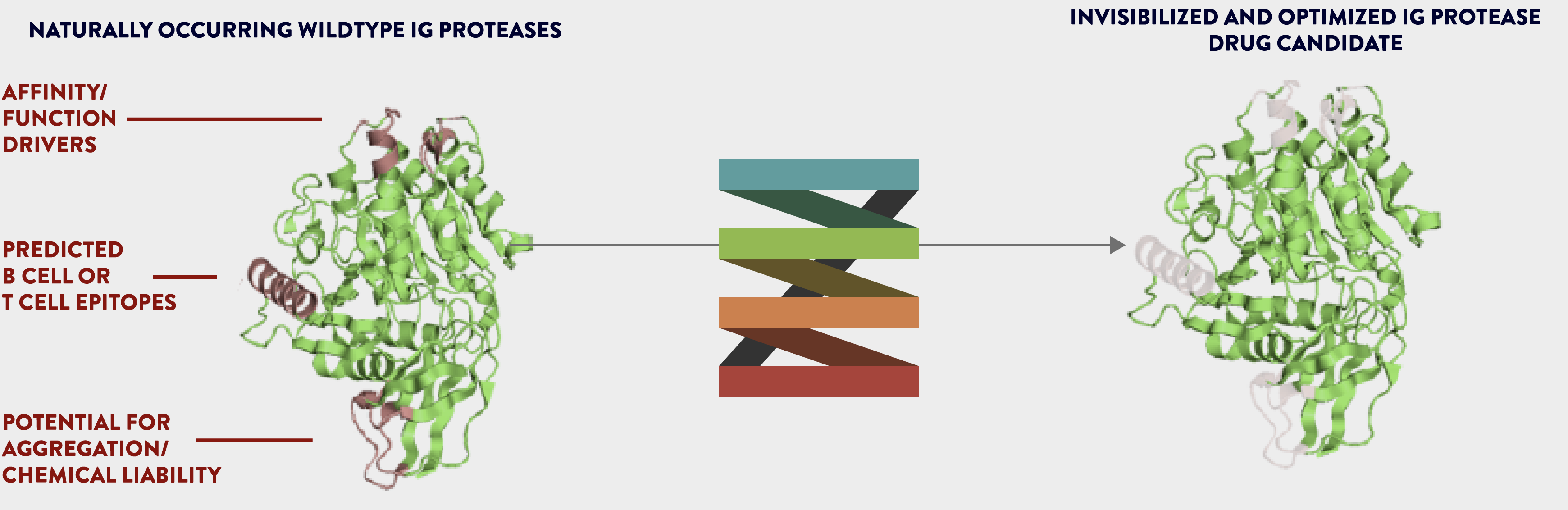
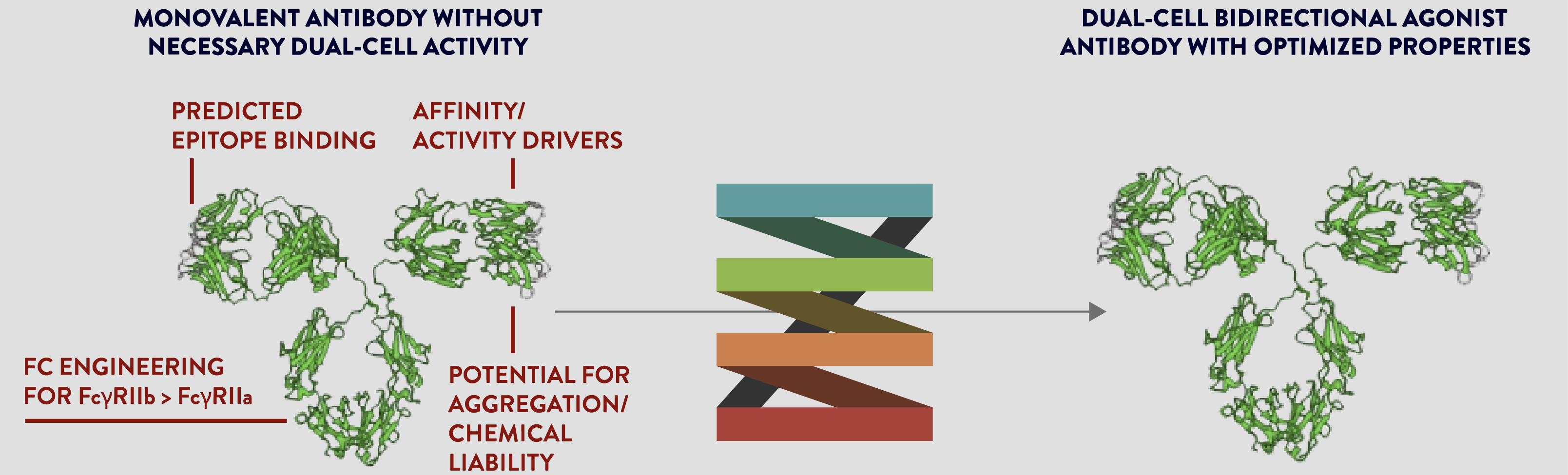
We generate novel biologic drug candidates on an unprecedented scale with optimized biologic activity and minimized immunogenicity .
What We Do
Our skilled team utilizes a variety of analytical techniques, such as x-ray crystallography and cryoEM to view proteins in 3D.
Where It Leads
3D models fuel our machine learning approaches and elucidate the assembly, function and interactions of our novel biologic drug candidate.
What We Do
Our proven team discovers antibody moieties and engineers natural proteins to create directed and optimized biologics using amino acid mutagenesis within protein sequences along with investigating and optimizing drug-like properties of biologics.
Where It Leads
We achieve improved functional activity, enhanced drug-like properties and good manufacturing attributes of our novel biologic drug candidates.
What We Do
Our experienced team investigates dysregulated immune mechanisms while effectively examining preclinical and potential clinical drug properties in novel in vitro human systems, and in vivo.
Where It Leads
We develope meaningful and precise therapeutics to alleviate dysregulated immune function in patients with autoimmune and inflammatory diseases.