Seismic’s DcB antibody approach targets dysregulated cell-mediated immunity by optimally engaging both T cells and antigen presenting cells, such as B cells, to restore immune homeostasis. Activating the normal inhibitory pathways of the immune system may control multiple diseases, such as multiple sclerosis, lupus and rheumatoid arthritis. DcB antibodies simultaneously engage multiple inhibitory pathways in more than one immune cell type thereby targeting and regulating both sides of the immune cell synapse.
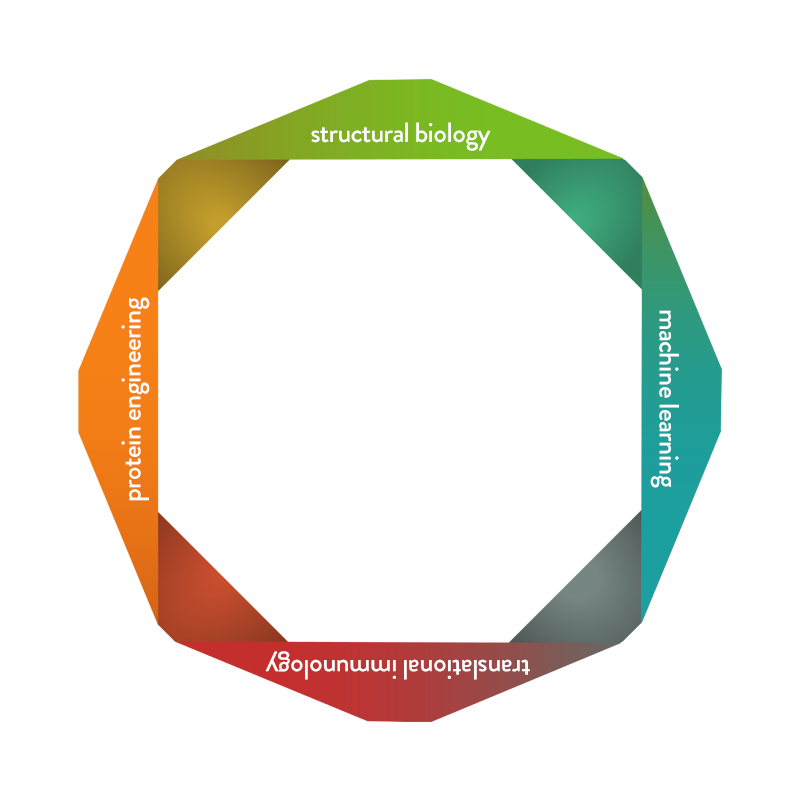
Our proven team of integrated protein engineers and immunologists work collectively with our cutting-edge ML team to optimize the drug-like properties of antibodies discovered through traditional discovery campaigns. Our IMPACT platform is critical to advancing the design of our FcγRIIb selective Fc domains by predicting mutations that tune for the desired binding properties at the Fc/FcgRIIb interface, informing optimal mutations to be tested experimentally for enhanced checkpoint agonism, ensuring invisibilization of these biologics to reduce the risk of immunogenicity in the clinic, and predicting antibody epitopes to support epitope binning and epitope/function relationship analysis.

How we optimized our DCB Antibody +
- Map protein-protein interfaces using machine learning thereby enabling design of FcγRIIb selective Fc domains
- Map protein-protein interfaces using ML thereby enabling design of FcgRIIb selective Fc domains
- Predict epitopes recognized by our proprietary antibodies to enable in silico epitope binning and epitope/function relationship elucidation
- Enable de novo paratope generation in silico for antibody generation/discovery
- Optimize developability of antibodies
- Map and remove immunogenic T-cell epitopes to create novel inhibitory receptor agonist antibodies with increased invisibility